Introduction¶
Cytotoxicity is an ability of chemical compounds to destroy cells either directly or indirectly. Cytotoxicity assays are used in drug discovery to screen potentially toxic compounds. The results are routinely presented as percentages of affected cells vs concentrations used. These data are usually discrete or even disjoint, and do not even rely on the concept of drug dose (concentration per number of cells). Unfortunately, there are no suitable measure indices to estimate cytotoxic potential of a compound within a dose range.
|
|
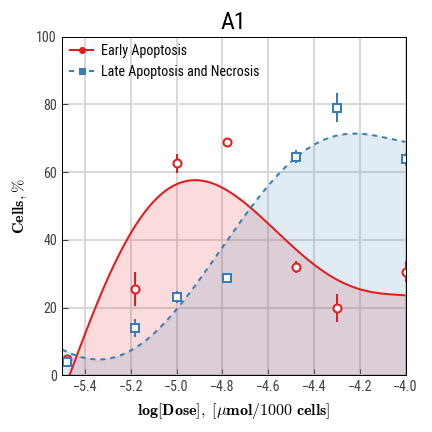
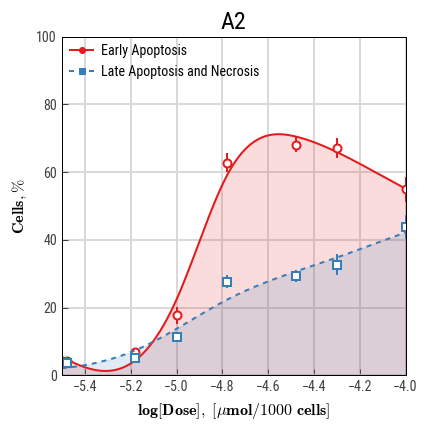
We have recently introduced a concept of cytotoxic power, a parameter that is based on numerical integration and drug doses instead of concentrations (Goncharov et al., 2018). Previously, we endorsed two other indices that may be used to highlight drug specifics (e.g. apoptosis prevalence over necrosis): apoptotic and necrotic difference (AND) and apoptotic and necrotic areas under curves (ANAUCs) (Smetanin et al., 2016, Galenko et al., 2017).
where x is a compound dose, f(x) is a dose-response function, Δt is incubation duration, a is minimal dose, and b is maximum dose.
This Python package is a proof-of-concept implementation of cytotoxic power calculation algorithm to assist researchers in processing cell assay results and presenting data.